- Seaborn - Home
- Seaborn - Introduction
- Seaborn - Environment Setup
- Importing Datasets and Libraries
- Seaborn - Figure Aesthetic
- Seaborn- Color Palette
- Seaborn - Histogram
- Seaborn - Kernel Density Estimates
- Visualizing Pairwise Relationship
- Seaborn - Plotting Categorical Data
- Distribution of Observations
- Seaborn - Statistical Estimation
- Seaborn - Plotting Wide Form Data
- Multi Panel Categorical Plots
- Seaborn - Linear Relationships
- Seaborn - Facet Grid
- Seaborn - Pair Grid
- Seaborn Useful Resources
- Seaborn - Quick Guide
- Seaborn - cheatsheet
- Seaborn - Useful Resources
- Seaborn - Discussion
Seaborn.pairplot() method
The Seaborn.pairplot() method is used to plot pairwise relationships in a dataset. Each numeric variable in the data will be spread over the y-axes across a single row and the x-axes across a single column by default, according to the axes grid created by this function.
A univariate distribution plot is created to display the marginal distribution of the data in each column for the diagonal plots, which are handled differently.
Additionally, you can display only a portion of the data or plot other variables in distinct rows and columns. This high-level PairGrid interface is designed to make it simple to create a few popular styles.
Syntax
Following is the syntax of the seaborn.pairplot() method −
seaborn.pairplot(data, *, hue=None, hue_order=None, palette=None, vars=None, x_vars=None, y_vars=None, kind='scatter', diag_kind='auto', markers=None, height=2.5, aspect=1, corner=False, dropna=False, plot_kws=None, diag_kws=None, grid_kws=None, size=None)
Parameters
Some of the parameters of the pairplot() method are discussed below.
| S.No | Name and Description |
|---|---|
| 1 | Data Takes dataframe where each column is a variable and each row is an observation. |
| 2 | hue variables that specify portions of the data that will be displayed on certain grid facets. To regulate the order of levels for this variable, refer to the var order parameters. |
| 3 | Kind Takes values from{scatter, kde, hist, reg}and the kind of plot to make is determined. |
| 4 | Diag_kind Takes values from{auto, hist, kde, None} and the kind of plot for the diagonal subplots is determined if used. If auto, choose based on whether or not hue is used. |
| 5 | Height Takes a scalar value and determines the height of the facet. |
| 6 | Aspect Takes scalar value and Aspect ratio of each facet, so that aspect * height gives the width of each facet in inches. |
| 7 | corner Takes Boolean value and If True, dont add axes to the upper (off-diagonal) triangle of the grid, making this a corner plot. |
| 8 | hue_order Takes lists as input and Order for the levels of the faceting variables is determined by this order. |
let us load the seaborn library and the dataset before moving on to the developing the plots.
Loading the seaborn library
To load or import the seaborn library the following line of code can be used.
Import seaborn as sns
Loading the dataset
in this article, we will make use of the exercise dataset inbuilt in the seaborn library. the following command is used to load the dataset.
exercise =sns.load_dataset("exercise")
The below mentioned command is used to view the first 5 rows in the dataset. This enables us to understand what variables can be used to plot a graph.
exercise.head()
The below is the output for the above piece of code.
index,Unnamed: 0,id,diet,pulse,time,kind 0,0,1,low fat,85,1 min,rest 1,1,1,low fat,85,15 min,rest 2,2,1,low fat,88,30 min,rest 3,3,2,low fat,90,1 min,rest 4,4,2,low fat,92,15 min,rest
Now that we have loaded the dataset, we will explore a few examples.
Example 1
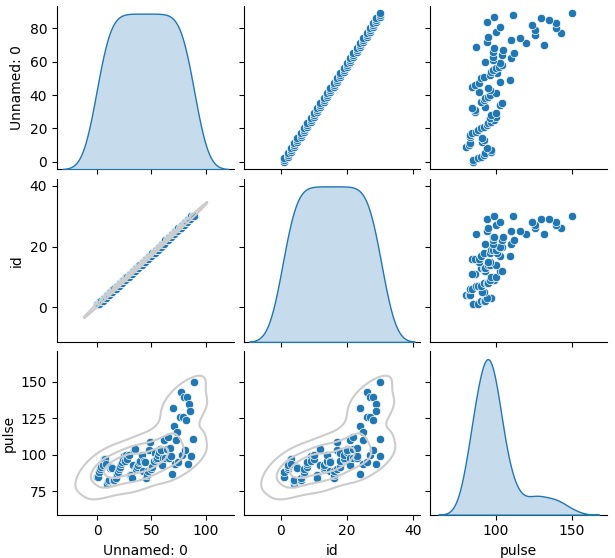
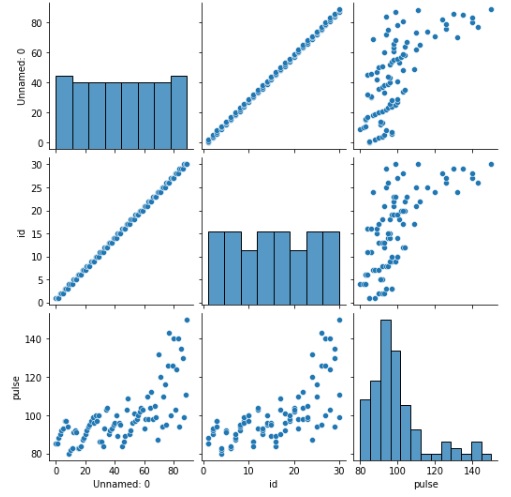
We will begin by calling the pairplot() method and passing the dataframe to it. This will yield multiple plots as can be seen below.
import seaborn as sns
import matplotlib.pyplot as plt
exercise =sns.load_dataset("exercise")
exercise.head()
sns.pairplot(exercise)
plt.show()
Output
Example 2
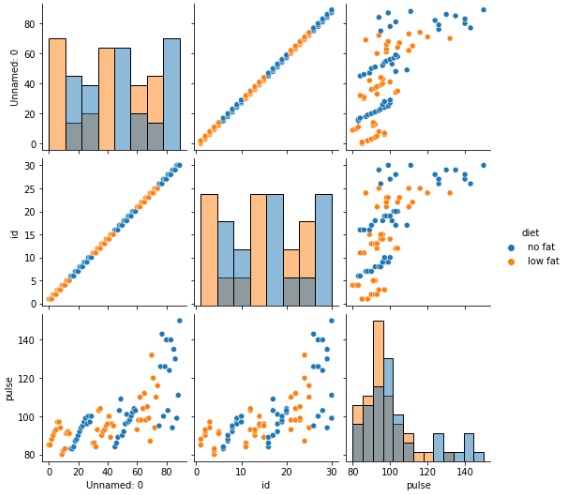
in this example, we will pass a few more parameters along with the dataframe to the pairplto() method. Since we are using the exercise dataset, we will use the diet categorical column and pass it to the hue dataset and we will plot graphs that are histograms in nature which is why the diag_kind a parameter is passed a value of hist.
import seaborn as sns
import matplotlib.pyplot as plt
exercise =sns.load_dataset("exercise")
exercise.head()
sns.pairplot(exercise, hue="diet", diag_kind="hist")
plt.show()
Output
The output obtained is seen below,
Example 3
We will understand the usage of map_lower() function in this example. We will save the pairplot into a variable g and the map_lower function id then applied to it. So basically, for every unique hue value that is present the func that is given to map.lower will be run (for each variable). When no hue is given the function will only be run once on all the relevant data (for each variable).
import seaborn as sns
import matplotlib.pyplot as plt
exercise =sns.load_dataset("exercise")
exercise.head()
g = sns.pairplot(exercise, diag_kind="kde")
g.map_lower(sns.kdeplot, levels=4, color=".8")
plt.show()
Output
The output obtained is as follows −